Downloading a static copy of the AACT database with Python
Hi, in this post I will share the method I used to download a static copy of the "Access to Aggregate Content of ClinicalTrials.gov" (AACT) database.
Before starting, a good question to ask is why would you want to download this information? As a health care professional or a data professional working in the health care realm, having access to a comprehensive database of clinical trials can be very useful. In particular, the AACT database is one of most comprehensive and detailed databases of clinical trials I have found: it is updated every day, it has good documentation and is really easy to use.
To know a bit more about the information it contains y recommend you visit the AACT database website and the ClinicalTrials.gov website. Here is a schematic of where the data comes from:

Other presentations of the AACT database
Besides downloading static copies of the database, you can also access the AACT database through a web interface, or a SQL database. You can find more information about these options in the AACT database download section.
Requirements
-
A working linux installation. I'm running Ubuntu 22.04 in WSL (Windows Subsystem for Linux) on Windows 11.
-
A Python environment. I'm using miniconda with a Python 3.11.7 virtual environment.
-
The
requestspython package. You can install it with the following command (remember to activate your virtual environment before running the install command):
pip install requests
Download the database
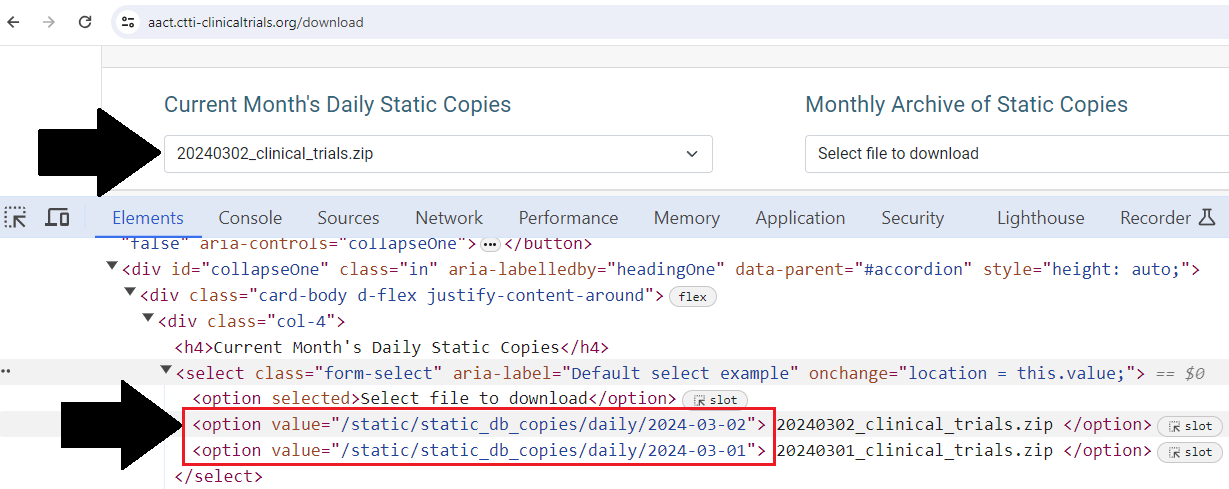
The first step is finding the URL from where we can download de database. From the dropdown menu, we can see that each option tag is linked to the url.
/static/static_db_copies/daily/<date>
With that information we can recreate the download URL. Check the URL variable in line 34 of the code below.
Here is the complete code, feel free to read the comments to understand what each part does and copy the code to your own project:
| download-database.py | |
|---|---|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 | |
To run the python script you can use the following command:
python download-database.py
After a couple of minutes, the script should finish and you should have the following directory structure:
.
├── data
│ └── 2024-03-01_aact_database
│ ├── data_dictionary.csv
│ ├── nlm_protocol_definitions.html
│ ├── nlm_results_definitions.html
│ ├── postgres.dmp
│ └── schema.png
├── downloads
│ └── 2024-03-01_aact_database.zip
└── download-database.py
The file we are interested in is the postgres.dmp file, which is a PostgreSQL database dump (backup) of the AACT database. We can use it to recreate a local copy of the database.
The other files are metadata, which I didn't find very useful, I prefer to use the information in AACT website to get some insight of how the database is structured.
Conclusion
In this post I shared a method to download a static copy of the AACT database using the python requests library. In upcoming blog posts I will show how to download the database metadata and how to create a local database with the downloaded dump file and docker.
If you find this content useful, please consider supporting my work by buying me a coffee.